The vast and growing number of publications in all disciplines of science cannot be comprehended by a single human researcher. As a consequence, researchers have to specialize in narrow subdisciplines, which makes it challenging to uncover scientific connections beyond their own field of research.
The Problem
As an interdisciplinary scientist even finding the right word to search for is not easy if you are unfamiliar with a new topic or field. When reading a paper, searching for relevant sections using CTRL+F is too dumb. Searching for "feeding" won't show potentially relevant sections like "nutrition".
The Goal
A smart search engine that facilitates finding semantically similar terms and reduces search time instead of guessing terms for CTRL+F
The Team
Without the help of my genious friends and lots of free time this idea would have not been possible
- Joanna Kaczanowska: Problem description
- Christoph Götz: Conception, Scrapy, data-mangling and Gensim model
- James Dommisse: AWS EC2 backend and Flask frontend magic
- Johannes Schuh: Search visualization Javascript magic
The Result
- Used Scrapy to download scientific papers from Pubmed et al
- 100k+ papers, 500M+ words from 2k+ journals
- Vectorized the topics using Gensim (word2vec)
- Search bubble in browser that takes search term and highlights semantically similar terms on the website you search in
- Biochemical/medicine terms supported only
Ok show me how this looks like
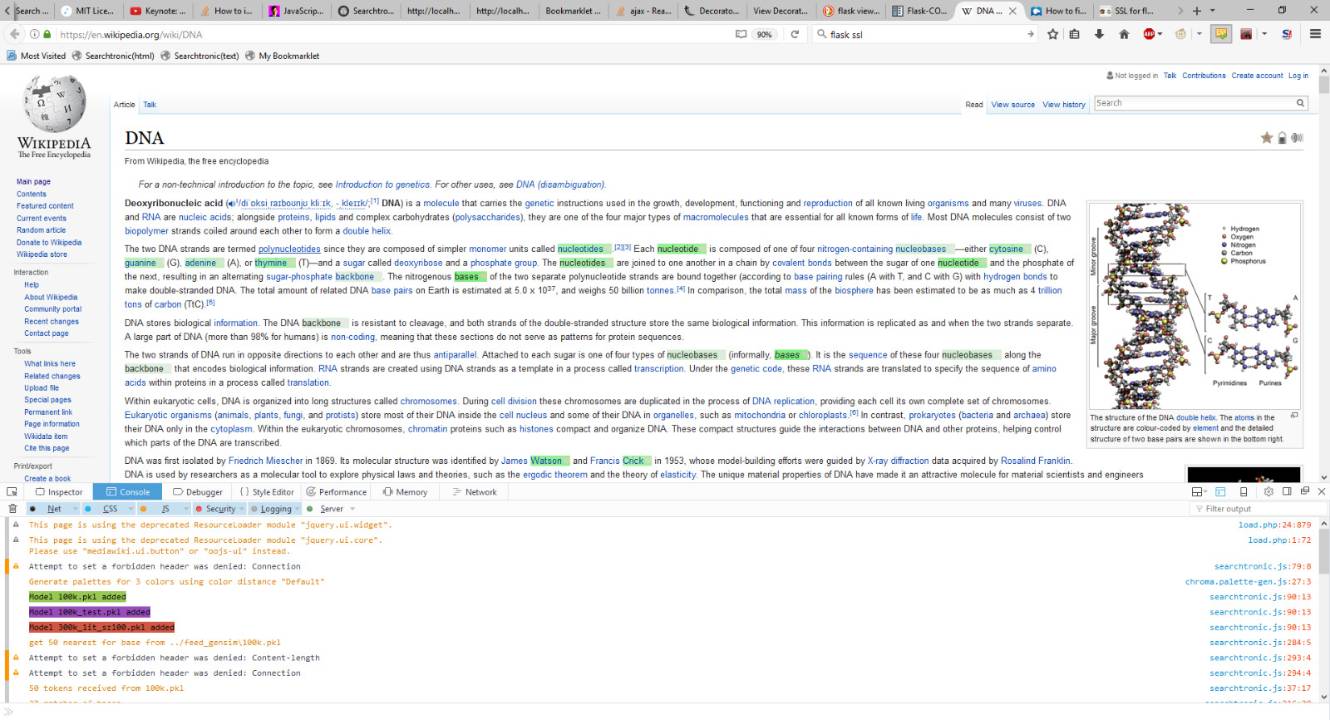
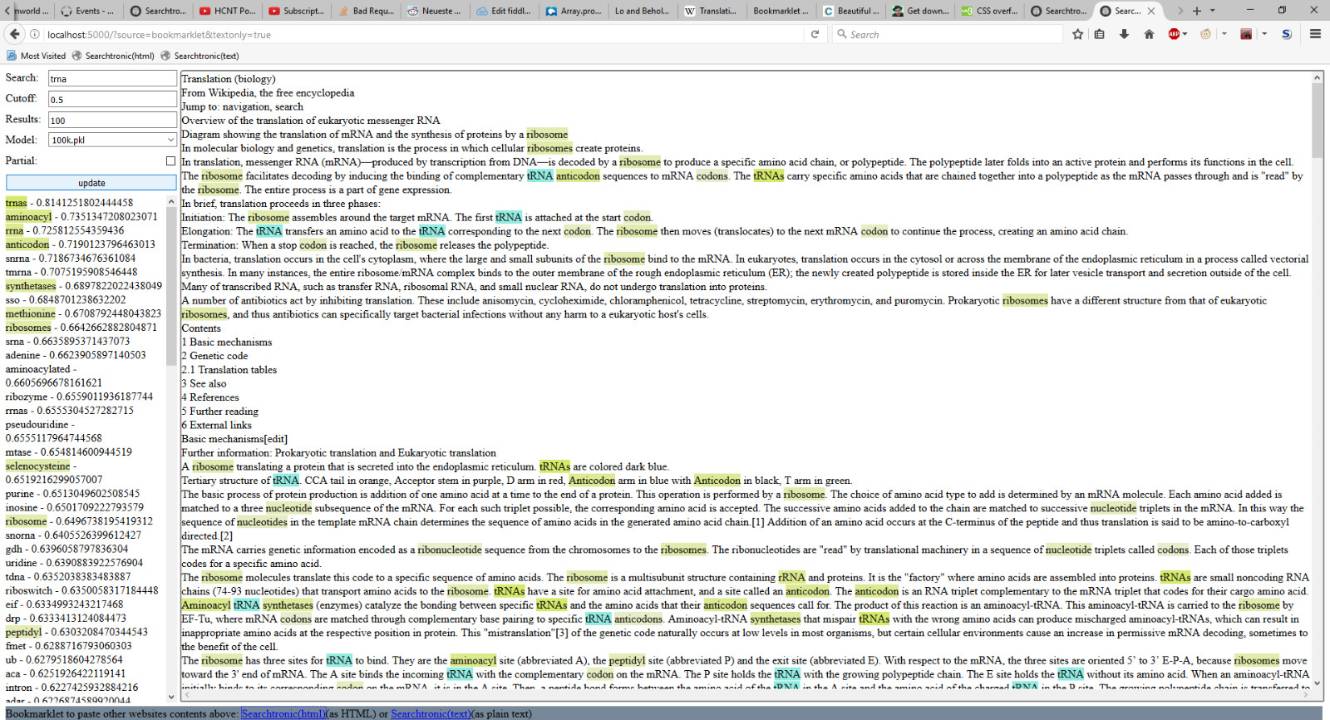
Below you see the user interface of the model in practice. You search for a term like "DNA" and magically terms like nucleotides, bases, cytosine, thymine, backbone and even Watson & Crick show up. Isn't that handy?
In less obvious circumstance this could have saved you the trouble of learning knowing what those terms mean and if they are relevant to your searchterm.
Sign up to the newsletter to get new posts straight to your inbox!
And here the backend
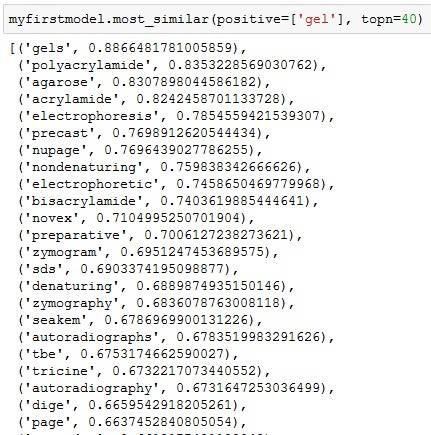
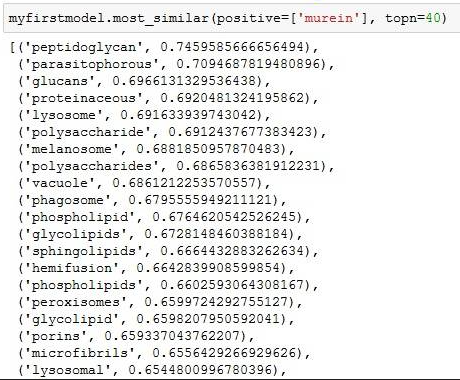
Searchterm is tRNA on the wiki article of Translation (RNA to protein). The numbers on the left is the "semantic distance" encoded in vector space. All you need to know is: 1 means similar, 0 means dissimilar. Documentation of Gensim explaining how it works is here.
The data
Every model is only as good as its data. Pubmed has an API which can download open-access papers in text-form. So this is what I drained over quite some time to not overburden their system which resulted in the following:
500M+ words from 2k+ journals and 100k+ scientific papers
What went into the training came mostly from a few journals, that contributed a lot of papers (over 20k papers for the top journal, see below). Nonetheless, the top 10 journals are sound and established and come from the biology and health sector, so all good.
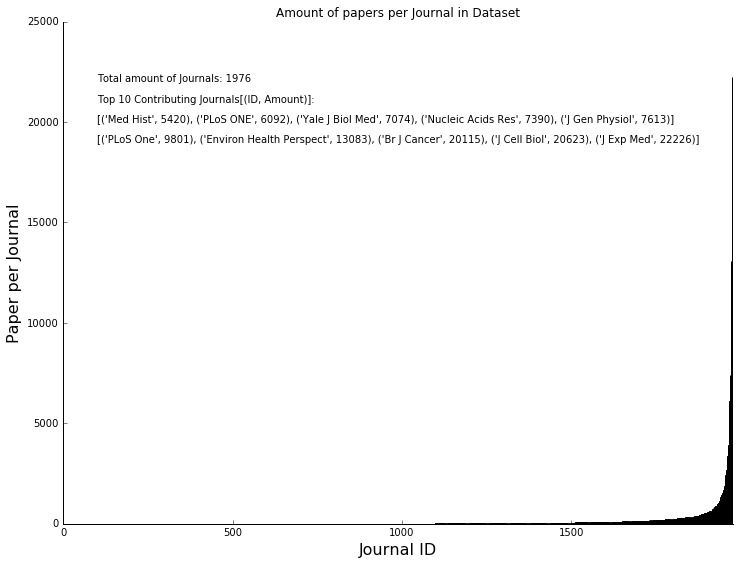
The distribution of sentences per journal also looks normal. Meaning we did not just download stubs but most papers contain 100s of sentences with only very few very short or very long article outliers.
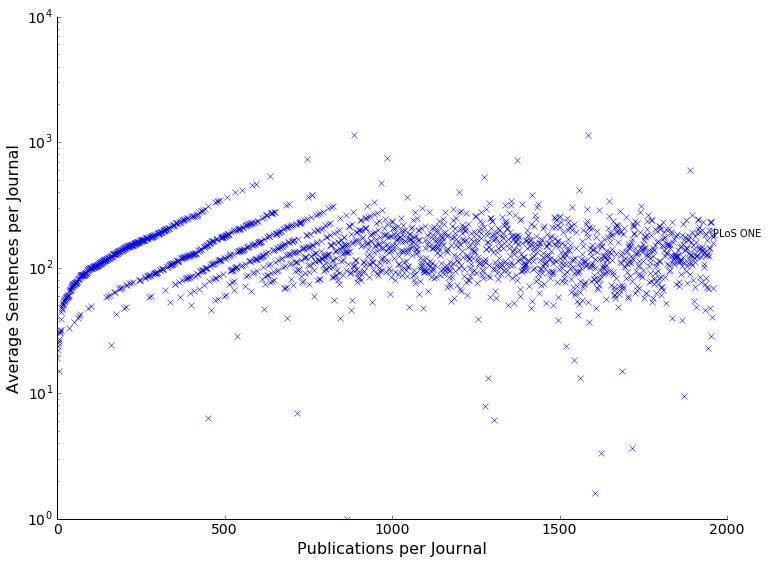
Looking at the word and sentence distribution alone shows that long articles with more than 10000 words or 1000 sentences are very seldom, respectively.
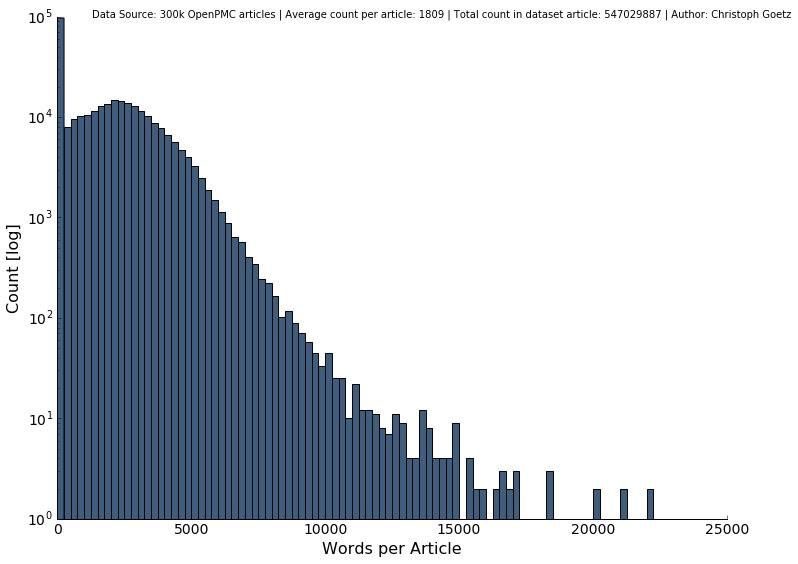
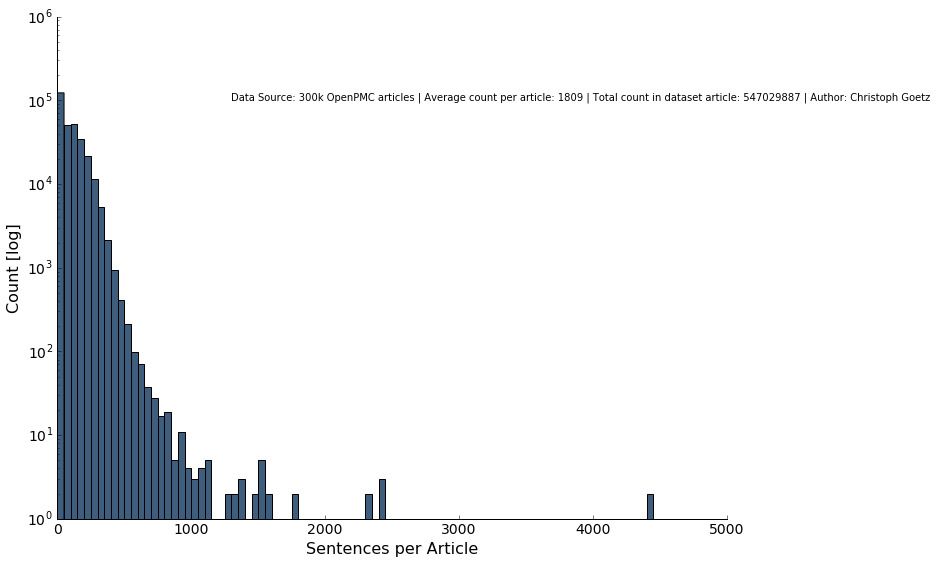
In total, we have 547,029,887 words from relevant journals from not too long and not too short papers of the medical field. This seems like a good dataset to start with.
Compared to Wikipedia with 4.1 billion words, this means the corpus contains the equivalent of 13% of Wikipedia, but highly specific to the medical and biological field.
Lets have some fun with it
The semantic search worked as shown in the UI above, but there must be more in the data to tinker with. Lets dig a bit, shall we?

Can we discover ultimate truths about the universe?
Can we learn something we already know from the data and the derived models? Could an alien that discovers after humanities climate-change induce collapse of civilization a hard-disk which contains this model and its knowledge to derive fundamental principles of nature without knowing what the words mean in the first place? Just because of the semantic similarities?

We can use a cool feat of vector-encoded information for this: Semantically encoded concepts can be investigated using simple mathematical operations like subtraction and addition. "King" - "man" + "female" = "Queen" actually works when trained on a normal text corpus (To take with a grain of salt).
Other examples to drive home the point:
- Paris - France + Germany = Berlin (for EU citizens)
- Irak + oil + weapons of mass destruction = Freedom (for US citizens)
- Leberkäsbrötchen - Fleisch + Milch = Käsebrötchen (For german speakers)
- Capitalism - Money + Beer = ?
The last example shows, that you can also discover things doing these kind of operations. Will it be heaven, hell or something completely different? Maybe a tech-savvy sociologist will answer this question for us. Lets stick to biology for now:
A well known dogma in biology is that information flows from DNA via transcription to RNA via translation to proteins. This should be well encoded in the literature as literally all of molecular biology relies on this to be true. Semantically this should be encoded as follows:
DNA + transcription = RNA
RNA + translation = Protein
So does the model actually produce the correct results if we do the subtraction? The below screenshot shows the following operations and should yield DNA and RNA as a result, respectively:
RNA - transcription = DNA (?)
Protein - translation = RNA (?)
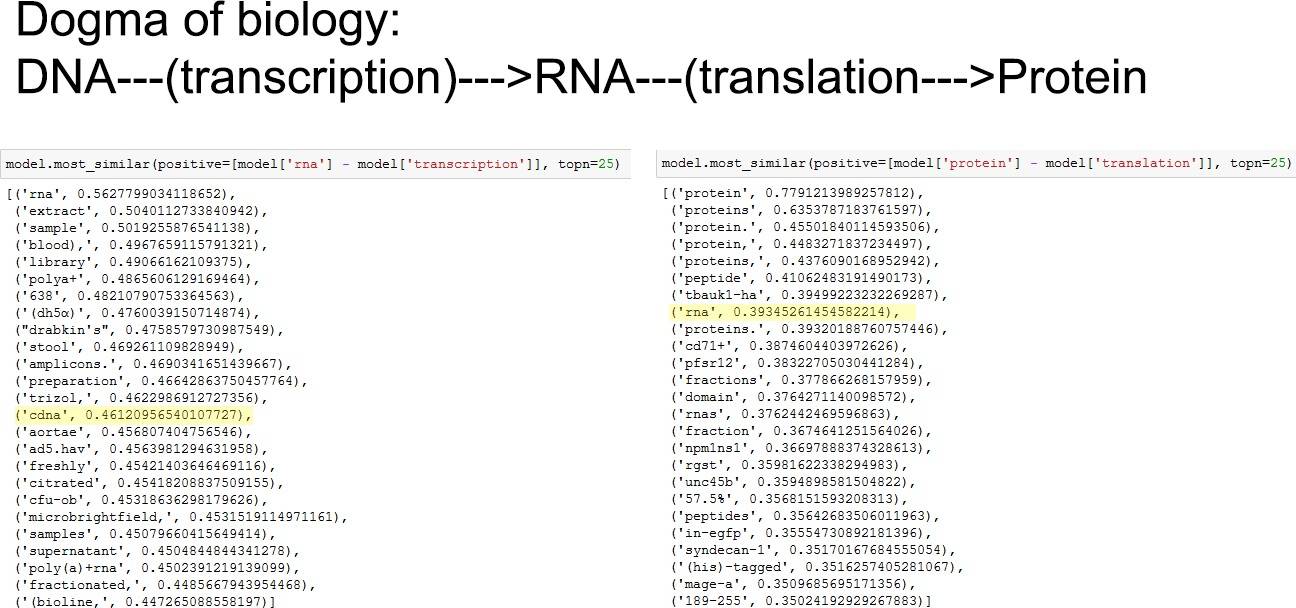
Amongst the top 25 hits it retrieves the correct outputs (highlighted in yellow).

Well. If you look closer its pretty difficult to rely on this to discover fundamental truths as there is lots of noise. One example is that the fundamentally important result "RNA" is almost equally important to "CD71+". According to Wiki: "CD71+ erythroid cells (CECs) appear in umbilical cord blood or peripheral blood of newborns". This is way too specific for such a general subtraction of "Protein-translation" and is likely a classic result of a badly trained model showing lots of unintended bias and brittleness. Strategies to overcome this would be:
- Careful data curation: Maybe the papers selected to train the model are too deep down in the weeds. One could filter the papers by impact prior to training to only include groundbreaking papers and not the vast majority of papers that just iteratively improve
- Regularization: Good old augmentation
- Switch technology: LSTMs are much more capable it turns out, see below
Influence of data source
Do more papers help improve the output quality or make it worse? Important biochemical information could be diluted in the myriad of medical case-report papers from across the globe [1].
Medical case reports are amazing though. Check out this one published in the British Journal of Obstetrics and Gynaecology involving Oral Sex, a Knife Fight and Then Sperm Still Impregnated Girl: https://blog.esciencecenter.nl/king-man-woman-king-9a7fd2935a85 ↩︎
I retrained the models with PLOS and the whole 2k journals (Model called 300k for whatever reason). Reason: PLOS is very good and focused while the other 1999 papers sometimes have shady reputation or are very fringe.

And now we learn another important lesson: if you have no metric on how to define success you have a hard time knowing what it actually means. Qualitatively it looks like PLOS seems to be ok, but has a few oddballs like "well-characterized". The model trained on the full corpus sounds very nerdy and does not show any obvious outliers which is good in this case.
Lessons Learned
My big takeaway was the intuition that knowledge exists inside of bubbles and knowledge creation is something like budding yeasts: Knowledge expands within the bubble, meets other bubbles, cross-talk between the bubbles ensues (sex) and a new bubble gets created containing information of both parent-bubbles. But let me explain first.

Limits of technology when generalizing
We started out with cell biology because lots of open data is available and I had an intuition for what makes sense. Can we actually generalize this to everything? For example semantic search in other domains of Chemistry, Engineering, Legal texts, Patents, etc?
Lets look at the below example of a concept like fat in two different contexts. The model as it is trained right now has no means of figuring out what's the correct context. This means that we dilute the semantic distances if we go to n-contexts. We will end up searching for fat and receiving all kind of answers that are true for all contexts it was trained on.
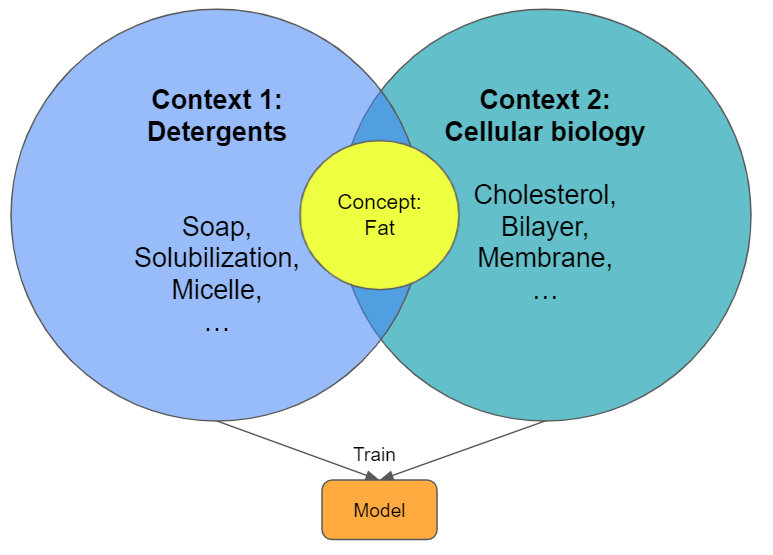
We are only interested in a model that works in our context. The text we search in provides the context and naturally acts as a "filter" of the search results. On a wiki-page of cell biology there is little talk about soaps and washing powders, so we should only find the relevant terms. Nonetheless, we don't make it easy for the model during training.
So why not train two different models for each context? Journals are actually a good proxy of context as they (sometimes) specialize on a certain aspect of a field. Lets take again the British Journal of Obstetrics and Gynaecology. What would a model trained only on this context be useful for? For people learning Gynaecology certainly a lot as its highly specialized. It might even be possible for people outside the field to discover completely new associations: What is semantically most relevant to Gynaecologists when you search for "fat"? Why does it matter? Should we care?
What you see here is a method of actually using this model to discover existing knowledge that is encoded but neither obvious to the people inside and outside of the field (Is it knowledge if it already exists but nobody knows about it?). But more to that part later.
Training two different models on two different contexts would look like below: Two models are trained, one for each context and then based on the context the "most appropriate" context is selected and used for inference. We need to add another onion-shell, namely a kind of classifier that shows us the right context to pick the most appropriate model and we are good to go.
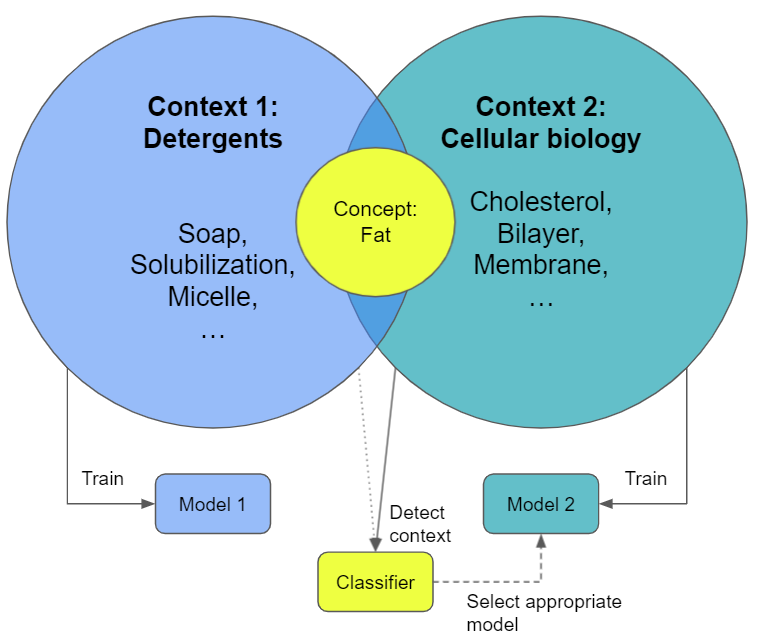
In theory this would greatly improve the prediction results within the correct context and make everyone happy, right? Getting rid of confusing labels from a different context makes the models happy and convergent. We would train a myriad of nerd-models orchestrated by the classifier that knows when to talk to what nerd.
However, in practice this is a terrible idea if we come to an n-context scenario: Where do you draw the borders between fields/contexts? What overlap is still tolerable? When do you start "budding-off" your new context? Can you automate that robustly? What if you add the context of diet, obesity, or fat beats/rhymes/dancemoves? What about a combination of them (e.g. an obese biochemist holding a piece of butter, laying down fat dancemoves on a soapy dancefloor)?
Figuring all of that out by yourself is feature engineering deluxe. You need another onion-shell of detecting the right context via a classifier which adds another element of uncertainty and prediction error to it.
Luckily humanity has solved this problem before and its called deep learning.
Neuronal networks are much better at layering contextual information while preserving local information [1] compared to the tech we used in 2016[2].
The unreasonable effectiveness of deep learning in artificial intelligence https://www.pnas.org/doi/10.1073/pnas.1907373117 ↩︎
Karpathy on the competing tech RNNs from 2015 http://karpathy.github.io/2015/05/21/rnn-effectiveness/ ↩︎
Thank god I work in image processing because you can actually look at things and have your visual cortex jump in the game to make sense of it. Language requires to purely juggling with pure information which makes it tricky at times. The good ol' Unet is certainly a good example of mixing both local information and context. Deep learning is unreasonable effective because of the depth that conserves the relationship between the local/concrete (word) and the abstract (context) information. In short, individual Pixels on top, meaning on the bottom, both nicely connected. So smart, Mr. Ronneberger already has 50k citations of his concept from 2015.
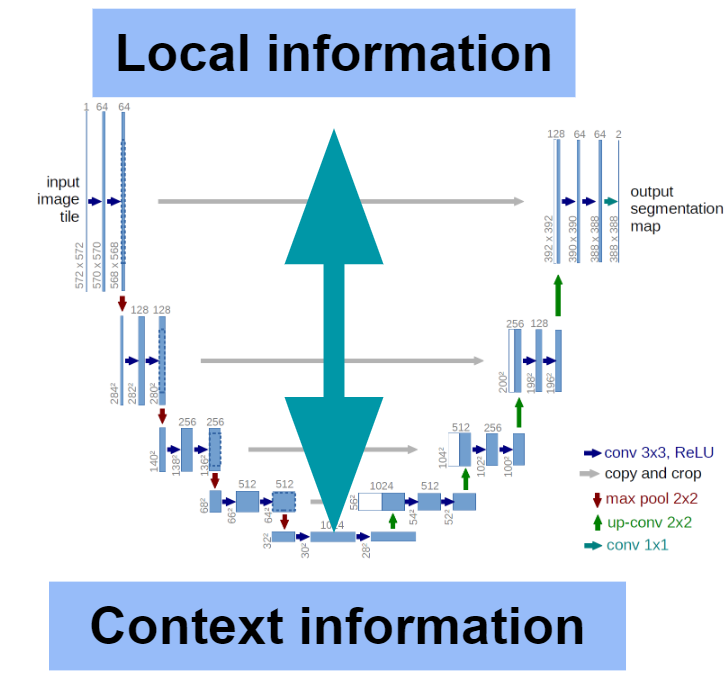
So while Gensims' vector embeddings have enough dimensions to encode local information, it lacks the contextual information encoding (based on my understanding from 2016).
Takeaway: Always stick to the state-of-the-art. This project was done in 2016ish, nowadays models with the like of openAIs GPT3 are taking care of all the problems described above.
The future of knowledge creation
Thinking of words in terms of vectors and semantic meaning as distance between vectors was a fun exercise. Meaning of scientific fields are formed by "bubbles" of words that partially overlap with other fields (think fat used in nutrition, detergents and organic chemistry in very different contexts). New evolving fields are budding off existing bubbles until they come completely distinct from its original semantic home. This process of knowledge creation was fascinating but - as always - not really novel and just the beginning:
While researching this topic I talked to researchers in the quantum physics group of Zeilinger who just won the nobel-price in 2022. Markus Krenn worked on an even cooler project namely predicting future research trends in a pretty similar manner. His work was finally published in PNAS in 2020.
Using network theoretical tools, we can suggest personalized, out-of-the-box ideas by identifying pairs of concepts, which have unique and extremal semantic network properties.
Thats such a cool concept! Predicting your future ideas based on the knowledge that is already somewhere around you. This substantiates the fact, that most big discoveries are often discovered by multiple people simulataneously (e.g. Evolution or Mendeleian inheritance). If the basic knowledge is available and people spend enough time and energy on connecting the dots, the dots will be connected.
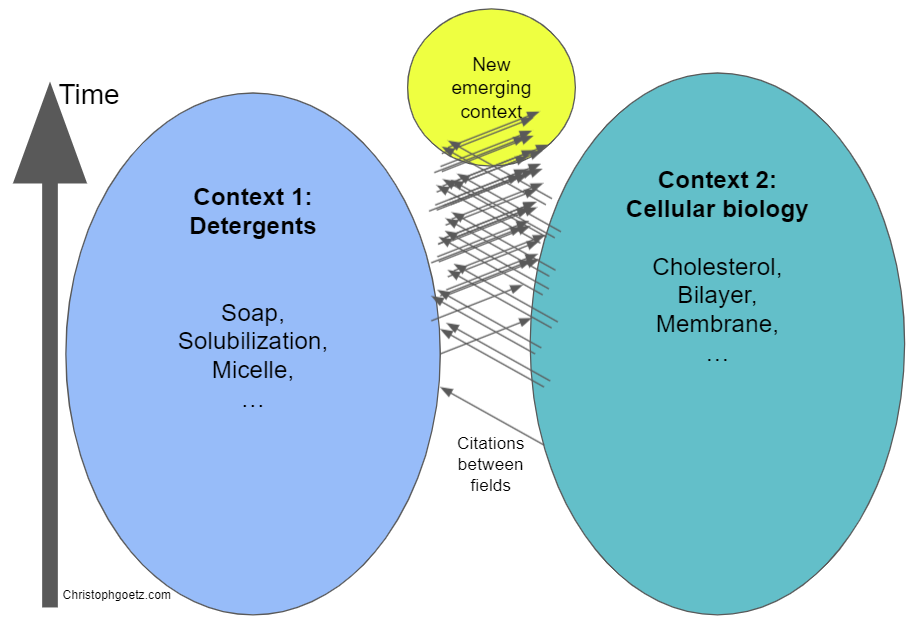
Citations can be seen as a force attracting two fields of research, cross-fertilizing and producing an offspring that contains information from both fields. I hope you now get the budding yeast analogy :D
Any real world applications?
This technology is currently used to predict trends in computer science or to determine the research priority for astronomy. Pretty cool stuff.
Conclusion
What started out as solving a problem for my (now) wife and tinkering with a simple tool for vectorizing words ended up in learning about fundamental vectors in cultural evolution. You never know what you don't know :D
Update Dec 2022
Facebook bought back then a paper-meta search tool with a pretty similar scope as this article. It was called "Meta" and after the Facebook rebrand we know why facebook bought the domain. Nonetheless, Yann LeCun, Facebook head of AI research continued developing tools tailored for the scientific community.
In Nov 2022 he launched Galactica.ai which was supposed to not only find similar words but directly write a whole scientific article on a topic.
A Large Language Model trained on scientific papers.
— Yann LeCun (@ylecun) November 15, 2022
Type a text and https://t.co/XKTkxs8Ae0 will generate a paper with relevant references, formulas, and everything.
Amazing work by @MetaAI / @paperswithcode https://t.co/IWGNAXiFeU
It was pulled off the web only three days later.
Galactica demo is off line for now.
— Yann LeCun (@ylecun) November 17, 2022
It's no longer possible to have some fun by casually misusing it.
Happy? https://t.co/K56r2LpvFD
Seems, science was and stays hard even with fancy new tools :D